dcHiC
dcHiC: Differential compartment analysis for Hi-C datasets
dcHiC HTML Demo Page
The following examples were created using dcHiC’s standalone visualization utility, which allows for single-command creation of HTML files for interactive compartment/genomic visualization. Examples from the paper are included below. Each file has a set of controls on the top, which allow users to search by region or by gene. These pages, available for download in the GitHub and created using IGV, will take a few seconds to load.
ESC vs NPC (100kb)
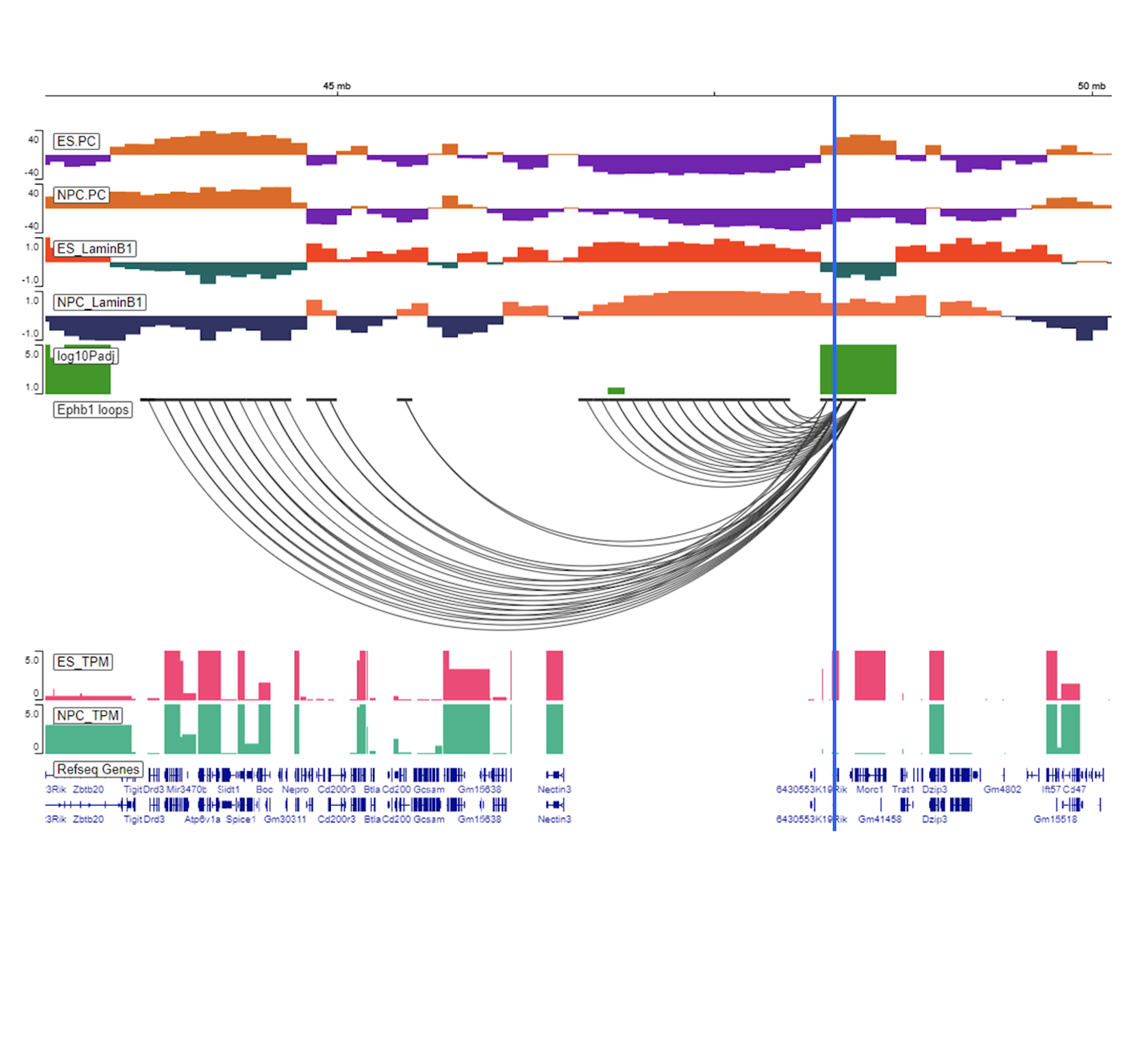
See this page for a comparison of ES and NPC compartments at 100kb. It contains the following tracks:
ES/NPC compartments
Differential Compartment Scores
Differential Loops
LaminB1 Data
RNA-Seq Data (TPM)
HOMER Differential Scores (for comparison)
ESC vs NPC (10kb)
See this page for a comparison of ES and NPC compartments at 10kb. It contains compartments and differential compartment scores.
ESC vs NPC vs CN (100kb)
See this page for a complete mice neural differentiation overview. It contains the following tracks:
ES/NPC compartments
Differential Compartment Scores
Differential Loops
LaminB1 Data
RNA-Seq Data (TPM)
Notable Regions
In all three ESC vs NPC examples, notable mESC-specific genes include: Dppa2/4 and Zpf42. Notable NPC/CN-specific genes include: Ptn and Ephb1.
Hematopoiesis
See this page for compartment dynamics during mice hematopoetic differentiation. This page contains compartments and differential compartment scores.
Notable changes include Meis1, Runx2, Sox6, and Abca13.
Lympoblastoid Data
See this page for compartment scores across 20 lymphoblastoid cell lines, using data from Gorkin et al, 2019. This page contains compartments and differential compartment scores.
Notable differential regions include NR2F2 and PTPRK.
Pseudo-Bulk scHi-C Data
Using postnatal neuronal scHi-C data, we created pseudo-bulk maps for analysis. In particular, Tan. et al created scHi-C maps for the cortex and hippocampus at days 1, 7, 28, 56, 309, and 347. We separated these into early (1/7), middle (28/56), and late (309/347) stage maps for each cell type – six different clusters in total.
See this page for the hippocampus comparison and this one for the cortex comparison.